About the Docking@Home Science: Advanced Concepts
|
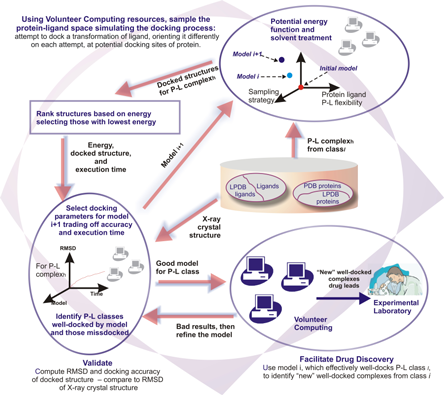
Docking@Home is part of the DAPLDS (Dynamically Adaptive Protein Ligand Docking System) project. This project aims to build a computational environment to assist scientists in understanding the atomic details of protein-ligand interactions. High-throughput, protein-ligand docking simulations are performed on a computational environment that deploys a large number of volunteer computers (donated compute cycles) connected to the Internet. The scales proposed in DAPLDS are not the traditional scales currently used in the life sciences. We deal with computational rather than experimental multi-scales.
Our multi-scale approach comprises three spanning scales (dimensions) of docking assumptions: protein-ligand representation, solvent representation, and sampling strategy. Within a scale, different scale values require different models and different algorithms to represent the models.In such a scenario, the two most critical challenges in dealing with multiple scales computationally are:
-
the ability to model biological systems with algorithms that dynamically adapt to the most appropriate value of each scale
-
the ability to assure that the algorithms can, indeed, be executed in the “required” amount of time using large numbers of distributed volunteer computing systems.
The latter point refers to having the necessary computational resources (CPU cycles, memory, network, etc.). The nature of these challenges requires collaboration among computational biophysicists, computational scientists, computer scientists, and system architects. These challenges and our main achievements are presented in this poster.
Our Objectives
-
Explore the multi-scale nature of dynamic protocol model adaptations for protein-ligand docking
-
Develop methods and models that efficiently accommodate these computational adaptations in VC environments based on BOINC
-
Extend knowledge with respect to protein-ligand complexes and make this knowledge accessible to the scientific community via cyber-infrastructures
|
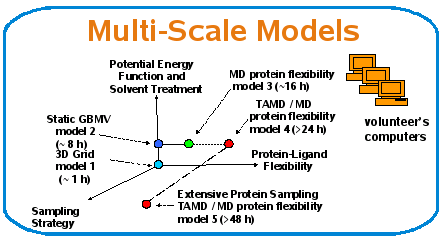
Multi-Scale Protein-Ligand Docking
-
Implement multi-scale docking models with different computational complexity and accuracy levels
-
Cluster protein-ligand complexes in classes based on characteristics
-
Define adaptive techniques to match models to classes dynamically
-
Matching based on quantitative values
Protein-Ligand Representation
-
Spanning scale from rigid to flexible representation of protein-ligand interactions
-
Coarse grid (1Å) with soft Lennard-Jones potential
-
Finer grid (0.25Å) with soft Lennard-Jones potential
-
All-atom representation of the protein-ligand interaction
-
Multiple protein structures of the same receptor considering small side-chain movements
-
Multiple protein structures of the same receptor considering large protein movements
Sampling Policy
-
Spanning scale from fixed to adaptive sampling of the protein-ligand docking space
-
Fixed number of trials per attempt and for each trial a fixed number of orientations per conformation
-
Change the number of trials per attempt as well as the number of orientations per trial
-
Different lengths for the heating and cooling phases as well as minimization in MD simulation
Potential energy and Solvent Treatment
-
Spanning scale from less accurate to more accurate modeling of solvent treatment
-
Constant dielectric coefficient + potential energy
-
Distance-dependent dielectric coefficient + potential energy
-
Implicit representation of solvent using a Generalized Born model+free energy
Challenges in Computation
-
Implement robust docking simulations
-
Across heterogeneous, volatile and error prone machines
-
Explore adaptive scheduling policies
-
Need for reliable simulation environments for testing
-
Deal with different levels of resource availability and reliability
-
Prevent starving machines and reduce redundant computation
-
Implement multi-scale algorithmic adaptations
-
Accommodate adaptations in cyber-infrastructures
-
Characterize resources and docking systems+models
-
Design techniques for selection of models and resources at run-time
|
View Multi-Scale Models
View DAPLDS
|
Go back to the
Science Introduction
View
Papers
on the subject
Learn more about Docking@Home terminology in the
Glossary


